MIMIC-IV 重症医学数据库部署与使用
详细操作步骤可看这两篇文章:
MIMIC数据库安装保姆级教程(上)
MIMIC数据库安装保姆级教程(下)
安装常见问题(一)
安装常见问题(二)
MIMIC-IV数据库安装常见问题(三)
MIMIC-IV数据库安装常见问题(四)
简介
医疗数据库不仅是我们进行医疗大数据分析的主要来源,同时也是科学研究的重要基础,可以用来改善患者的治疗和诊断,帮助改善医疗服务的质量。它也可以支持临床研究和政策制定,帮助改善健康管理模式,提高治疗效果,减少治疗成本。
MIMIC(Medical Information Mart for Intensive Care)是一个由_麻省理工大学_联合_贝斯以色列女执事医疗中心_等共同建立的重症监护数据库,它是目前急诊和危重症领域最大、可以公开访问的临床数据库。MIMIC最新版本MIMIC-IV发布于2020年,其中最新的数据发布于2023年2月6日。
MIMIC-IV提供了一个多维度的数据集,可以支持重症医学领域的研究,它包含来自美国麻省总医院的多种类型的重症患者的数据。
尽管MIMIC是公开数据集,但是该数据集的下载需要通过考核与人工审核。
https://physionet.org/content/mimiciv/2.2/

内容
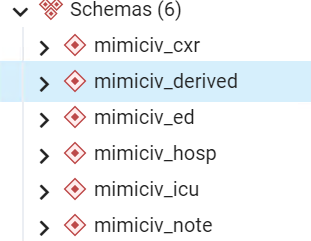
截至本文发布时的最新版本(v2.2)时,MIMIC IV总共公布了HOSP、ICU、ED、CXR、NOTE五大模块的数据。
(目前还有waveforms待发布
部署
本文将以Windows为例,将MIMIC-iv导入PostgreSQL数据库中。
下载安装PostgreSQL
https://www.enterprisedb.com/downloads/postgres-postgresql-downloads

下载数据
略,解压到F:\mimiciv
其中.sql.gz无需解压
Clone导入代码
|
|
HOSP+ICU模块
DROP DATABASE IF EXISTS mimiciv; CREATE DATABASE mimiciv OWNER postgres;
\c mimiciv;
\encoding 'UTF8'
\i F:/mimic-code/mimic-iv/buildmimic/postgres/create.sql; -- 创建表
\set ON_ERROR_STOP 1
\set mimic_data_dir 'F:/mimiciv' --定义数据目录
\i F:/mimic-code/mimic-iv/buildmimic/postgres/load_7z.sql; -- 导入数据
\i F:/mimic-code/mimic-iv/buildmimic/postgres/index.sql; -- 导入索引
\i F:/mimic-code/mimic-iv/buildmimic/postgres/constraint.sql; -- 导入约束
\i F:/mimic-code/mimic-iv/buildmimic/postgres/validate.sql; -- 验证导入
ED模块
基本同理
\c mimiciv;
\encoding 'UTF8'
\i F:/mimic-code/mimic-iv-ed/buildmimic/postgres/create.sql; -- 创建表
\set ON_ERROR_STOP 1
\set mimic_data_dir 'F:/mimiciv/ed' --定义数据目录
\i F:/mimic-code/mimic-iv-ed/buildmimic/postgres/load_7z.sql; -- 导入数据
\i F:/mimic-code/mimic-iv-ed/buildmimic/postgres/index.sql; -- 导入索引
\i F:/mimic-code/mimic-iv-ed/buildmimic/postgres/constraint.sql; -- 导入约束
Note模块
\c mimiciv;
\encoding 'UTF8'
\i F:/mimic-code/mimic-iv-note/buildmimic/postgres/create.sql; -- 创建表
\set ON_ERROR_STOP 1
\set mimic_data_dir 'F:/mimiciv/note' --定义数据目录
\i F:/mimic-code/mimic-iv-note/buildmimic/postgres/load_7z.sql; -- 导入数据
CXR模块
官方代码库中似乎没有导入SQL语句,我们可以手动创建
-- create.sql ------------------------------------------- -- Create the tables and MIMIC-IV schema -- -------------------------------------------
-- Creating schemas --
\echo '######################'
\echo 'Creating schemas.....'DROP SCHEMA IF EXISTS mimiciv_cxr CASCADE;
CREATE SCHEMA mimiciv_cxr;
-- Creating tables --
\echo 'Creating mimiciv_cxr tables.....'
DROP TABLE IF EXISTS mimiciv_cxr.chexpert;
CREATE TABLE mimiciv_cxr.chexpert
(
"subject_id" int8,
"study_id" int8,
"Atelectasis" varchar(255),
"Cardiomegaly" varchar(255),
"Consolidation" varchar(255),
"Edema" varchar(255),
"Enlarged Cardiomediastinum" varchar(255),
"Fracture" varchar(255),
"Lung Lesion" varchar(255),
"Lung Opacity" varchar(255),
"No Finding" varchar(255),
"Pleural Effusion" varchar(255),
"Pleural Other" varchar(255),
"Pneumonia" varchar(255),
"Pneumothorax" varchar(255),
"Support Devices" varchar(255)
);DROP TABLE IF EXISTS mimiciv_cxr.metadata;
CREATE TABLE mimiciv_cxr.metadata
(
"dicom_id" varchar(255),
"subject_id" int8,
"study_id" int8,
"PerformedProcedureStepDescription" varchar(255),
"ViewPosition" varchar(255),
"Rows" varchar(255),
"Columns" varchar(255),
"StudyDate" varchar(255),
"StudyTime" varchar(255),
"ProcedureCodeSequence_CodeMeaning" varchar(255),
"ViewCodeSequence_CodeMeaning" varchar(255),
"PatientOrientationCodeSequence_CodeMeaning" varchar(255)
);DROP TABLE IF EXISTS mimiciv_cxr.negbio;
CREATE TABLE mimiciv_cxr.negbio
(
"subject_id" int8,
"study_id" int8,
"Atelectasis" varchar(255),
"Cardiomegaly" varchar(255),
"Consolidation" varchar(255),
"Edema" varchar(255),
"Enlarged Cardiomediastinum" varchar(255),
"Fracture" varchar(255),
"Lung Lesion" varchar(255),
"Lung Opacity" varchar(255),
"No Finding" varchar(255),
"Pleural Effusion" varchar(255),
"Pleural Other" varchar(255),
"Pneumonia" varchar(255),
"Pneumothorax" varchar(255),
"Support Devices" varchar(255)
);
DROP TABLE IF EXISTS mimiciv_cxr.split;
CREATE TABLE mimiciv_cxr.split
(
"dicom_id" varchar(255),
"study_id" int8,
"subject_id" int8,
"split" varchar(255)
);
--load.sql ----------------------------------------- -- Load data into the MIMIC-IV schemas -- ------------------------------------------- To run from a terminal:
-- psql "dbname=<DBNAME> user=<USER>" -v mimic_data_dir=<PATH TO DATA DIR> -f load_gz.sql-- hosp schema
\cd :mimic_data_dir-- making sure that all tables are emtpy and correct encoding is defined -utf8-
SET CLIENT_ENCODING TO 'utf8';\echo '######################'
\echo 'Delete mimiciv_cxr table.....'
DELETE FROM mimiciv_cxr.split;
DELETE FROM mimiciv_cxr.negbio;
DELETE FROM mimiciv_cxr.metadata;
DELETE FROM mimiciv_cxr.chexpert;\echo '######################'
\echo 'Copying mimiciv_cxr table.....'
\echo 'Copying split.....'\COPY mimiciv_cxr.split FROM 'split.csv' DELIMITER ',' CSV HEADER NULL '';
\COPY mimiciv_cxr.negbio FROM 'negbio.csv' DELIMITER ',' CSV HEADER NULL '';
\COPY mimiciv_cxr.metadata FROM 'metadata.csv' DELIMITER ',' CSV HEADER NULL '';
\COPY mimiciv_cxr.chexpert FROM 'chexpert.csv' DELIMITER ',' CSV HEADER NULL '';
\echo 'mimiciv_cxr successfully generated.'
--index.sql -------------------------------------- -------------------------------------- -- Indexes for all MIMIC-IV modules -- -------------------------------------- --------------------------------------
-- cxr --
SET search_path TO mimiciv_cxr;
\echo 'Create mimiciv cxr index.....'
DROP INDEX IF EXISTS chexpert_idx01;
CREATE INDEX chexpert_idx01
ON chexpert (subject_id);DROP INDEX IF EXISTS chexpert_idx02;
CREATE INDEX chexpert_idx02
ON chexpert (study_id);DROP INDEX IF EXISTS metadata_idx01;
CREATE INDEX metadata_idx01
ON metadata (subject_id);DROP INDEX IF EXISTS metadata_idx02;
CREATE INDEX metadata_idx02
ON metadata (study_id);DROP INDEX IF EXISTS negbio_idx01;
CREATE INDEX negbio_idx01
ON negbio (subject_id);DROP INDEX IF EXISTS negbio_idx02;
CREATE INDEX negbio_idx02
ON negbio (study_id);DROP INDEX IF EXISTS split_idx01;
CREATE INDEX split_idx01
ON split (subject_id);
DROP INDEX IF EXISTS split_idx02;
CREATE INDEX split_idx02
ON split (study_id);
然后在控制台
\c mimiciv;
\encoding 'UTF8'
\i F:/mimic/cxr/create.sql; -- 创建表
\set ON_ERROR_STOP 1
\set mimic_data_dir 'F:/mimic/cxr' --定义数据目录
\i F:/mimic/cxr/load.sql; -- 导入数据
\i F:/mimic/cxr/index.sql;
导入结果